Transposable Element Dynamics
Transposable elements (TEs) are mobile genetic elements present in all domains of life that are able to change their position within the genome. There are two primary types of transposable elements that jump around in different ways. First are the transposons, which are the primary type of transposable element in bacteria and archaea. For example, transposons make up ~2-3% of E. coli's genome. For a transposon to jump, an enzyme, called a transposase, actually cuts the DNA that makes up the transposon out of the DNA. The transposase and excised DNA then find a new site in the genome, where the enzyme reintegrates the transposon DNA into the genome. Hence, transposons jump around through a "cut and paste" mechanism. The other primary type of transposable elements are the retrotransposons, which are the dominant type of transposable element in eukaryotes like humans. Rather than cutting themselves out of the genome, retrotransposons simply make copies of themselves by transcription into RNAs. These RNAs are then reverse transcribed and integrated elsewhere into the genome. Hence, retrotransposons jump around through a "copy and paste" mechanism. Because retrotransposons make copies of themselves, and those copies make copies, and so forth, they have exploded in number in higher organisms. The human genome, for example, is composed of ~45% retrotransposons, and "Junk DNA" is primarily composed of TEs. TE activity is the direct cause of many human diseases including muscular dystrophy and cancer, and the upregulation of TE activity in neural and tumor cells is thought to contribute to the plasticity and resilience of these tissues.
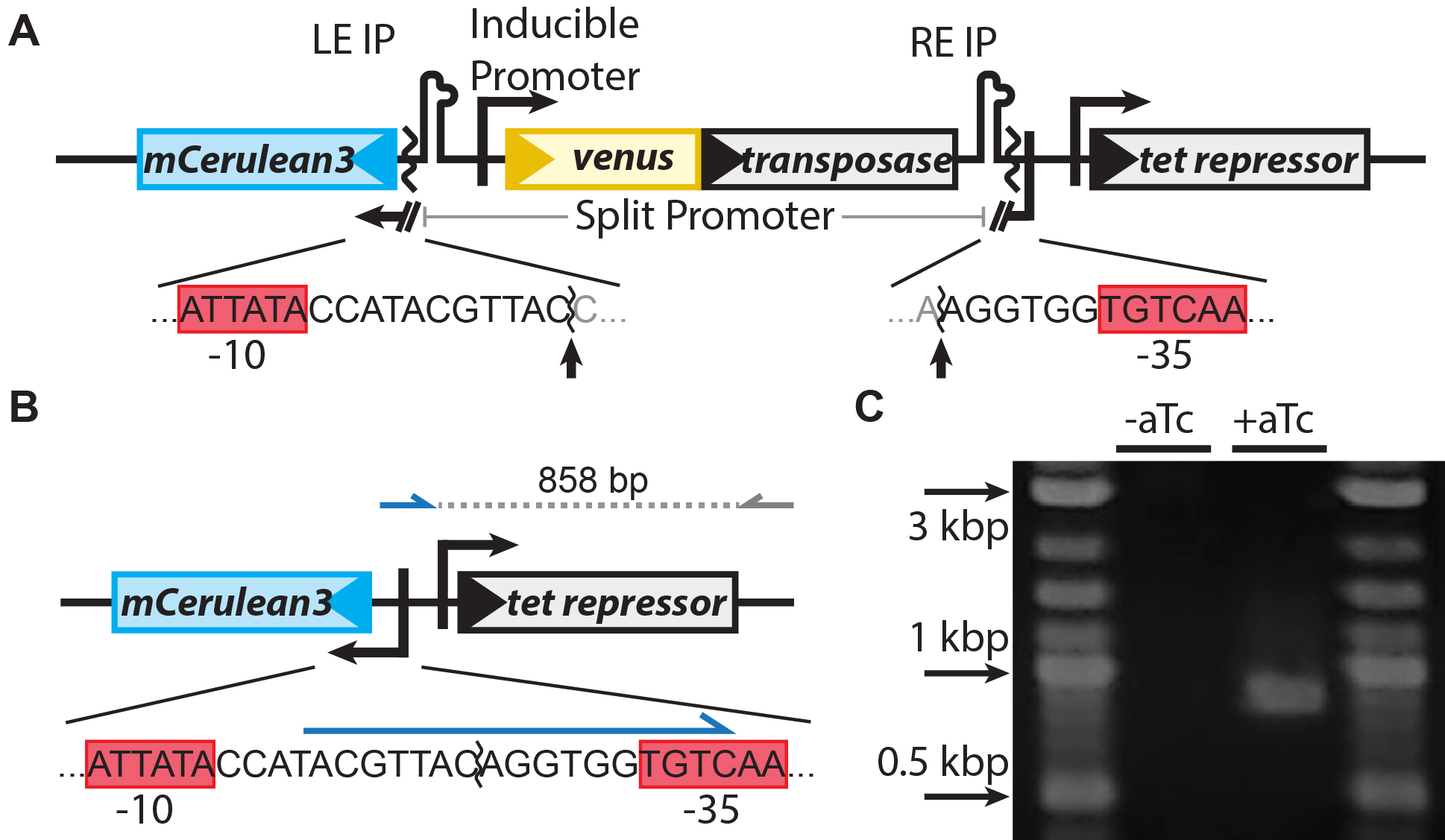
Figure 1: Our system for watching the the transposon IS608 jump around in living cells in real time. (A) Design of the system. A promoter for the blue fluorescent protein mCerulean3 is interrupted by the TE. The transposase, tnpA (gray), is inducible with aTc. Sequences of the interrupted promoter are shown below with -10 and -35 sequences highlighted by red boxes, cleavage sites indicated by arrows. (B) Upon excision, the promoter for mCerulean3 is reconstituted and the cell fluo-resces blue. A primer that binds the sequence formed after excision (blue arrow) was used to verify excision by PCR, generating an 858 bp amplicon. (C) PCR using these primers only generates product after induction.
Since transposable elements are so numerous and are such basic parts of the genomes of all living things, a complete understanding of their functions and activity could provide deep insights into many fundamental questions about evolution, development, and the etiology of human diseases. Unfortunately, current experimental techniques to study TEs have many severe limitations. For example, you might allow a population of cells to grow over time with some TE actively jumping around through their genome. After a while, you can then take a sample of cells and sequence them to see where in the genome the TEs have jumped. However, unless you are able to sequence single cells (which remains technically challenging), you will have to sequence with extraordinarily deep coverage to get a good measurement of all the transpositions in the sample. Furthermore, unless you take many samples over time, you also have no information about the dynamics of the process, or how transposition rates may change over time or with location in a clump of cells. Consequently, most existing methods generate data that are averages over collections of cells and over time, and provide no information about how rates may change from cell to cell, over time, or in different regions of space.
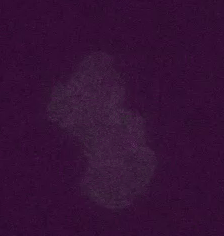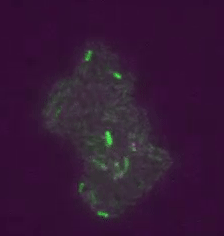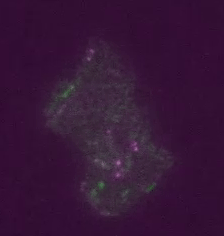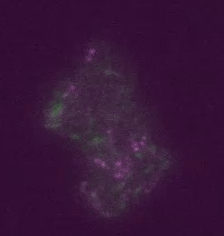
Figure 2: Transposons jumping around in live cells and in real time. Shown is a colony of ~400 E. coli cells. The reporter for transposon jumping is false-colored green, and the reporter for transposase levels is false-colored magenta. You can see that those cells who experience TE jumps (i.e. those cells that light up green) are also producing transposase (i.e. also light up magenta).
To remedy these limitations, we have developed techniques to directly visualize TE activity in individual cells and in real time (Figure 1). The idea is simple: when the TE starts jumping around, it makes the cell start producing a fluorescent protein; in this case the blue fluorescent protein mCerulean3. Hence, we can immediately detect those cells that have experienced TE activity simply by putting them under the microscope and watching them grow while looking for fluorescence. Moreover, we can also tag the proteins involved in transposition with other fluorescent colors; here, for example, we have fused the transposase to the yellow fluorescent protein Venus. We can then measure how much transposase is in the cell by how yellow they get, and measure the amount of resulting TE activity by how blue they get (Figure 2).
These data, recently published in PNAS [Kim et al., PNAS 2016], show that the activity rates of even the simplest synthetic TE fluctuate wildly over space and time, depending on the environment, the growth state of the cells, and their heriditary history. We are extending our measurements to study the activity of "wildtype" TEs, their activity and interactions within the chromosome, and their physiological effects on affected cells. Furthermore, we are further developing our systems to allow us to observe integrations of both transposons and retrotransposons in real time .